Understanding Orthology
Orthogroups, Orthologs, and Paralogs
-
• OrthogroupThe group of genes descended from a single gene in the last common ancestor of a group of species.
-
• OrthologsPairs of genes that descended from a single gene in the last common ancestor of two species.
-
• ParalogsPairs of genes descended from a gene duplication event.
Orthologs can be thought of as equivalent genes between two species, as they descended
from a single gene in the last common ancestor of that species. For example, the last
common ancestor of humans and mice is a small mammal which lived alongside the
dinosaurs. Individual genes present in that ancestor still exist in some form through their
descendants in both humans and mice, and those genes are orthologs.
Orthologs describe relationships between pairs of species, but we can extend this idea to larger groups of species. Humans, mice, and chickens share a common ancestor from a few hundred million years ago, before the dinosaurs had even emerged. We can describe a group of genes across all three species that were descended from a single gene in this ancestor - these genes form an orthogroup.
Look at the figure below, which shows data for three species: human, mouse and chicken.
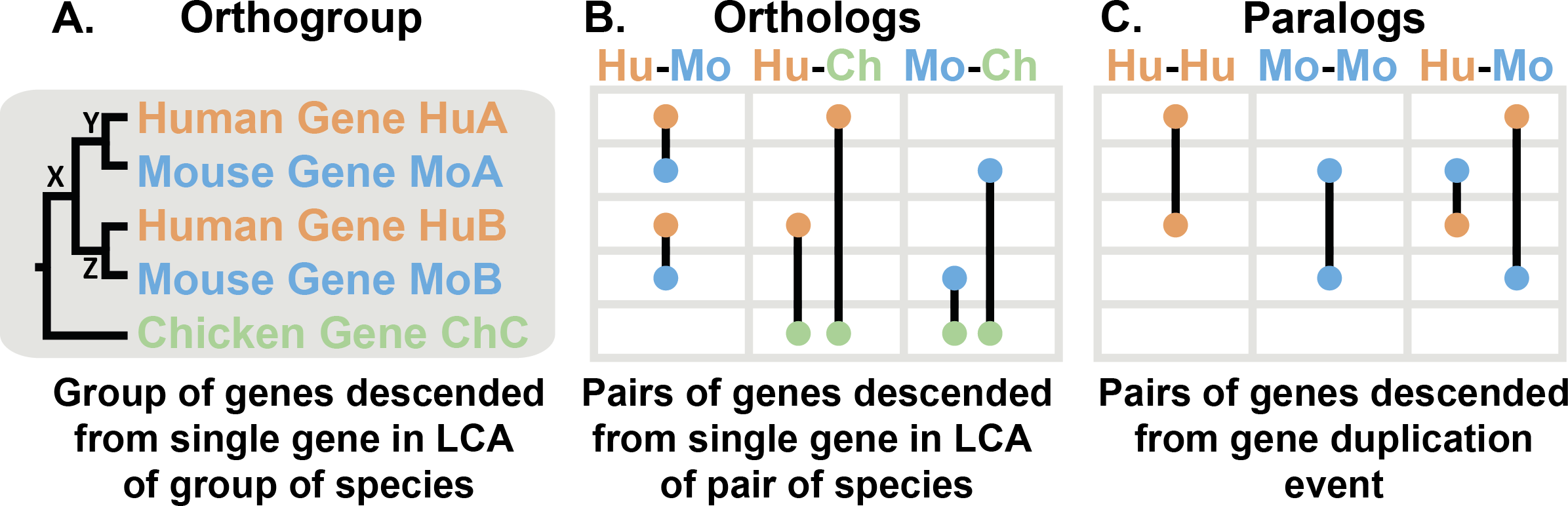
Figure 1: Orthologues, Orthogroups & Paralogues
The tree in Figure 1A shows the evolutionary history of a gene. First, there was a speciation event where the chicken lineage diverged from the human-mouse ancestor. In the human-mouse ancestor, there was a gene duplication event at X producing two copies of the gene in that ancestor, Y & Z. When human and mouse diverged they each inherited gene Y (becoming HuA & MoA) and gene Z (HuB & MoB). In general, we can identify a gene duplication event because it creates two copies of a gene in a species (e.g. HuA & HuB).
The mouse gene MoB is closer related to a human gene than it is to the other mouse gene MoA. This is because the gene duplication occurred in the ancestor, so each mouse gene is more closely related to its human ortholog (e.g. HuA and MoA), as they were both descended from a single gene. By contrast, HuA and MoB diverged at the gene duplication event. They aren’t descended from a single gene in the common ancestor of human and mice, so aren’t orthologs. Instead, they are paralogs (Figure 1C). Paralogs are more distantly related, they diverged at a gene duplication event in a common ancestor. Such a gene duplication event must have occurred further back in time than when the species diverged and so paralogs between a pair of species are always less closely related than orthologs between that pair of species. Paralogs are also possible within a species (e.g. HuA & HuB).
The chicken gene diverged from the other genes when the lineage leading to chicken split from the lineage leading to human and mouse. Therefore, the chicken gene ChC is an ortholog of HuA & HuB in human and an ortholog of MoA & MoB in mouse. Depending on what happend after the genes diverged, orthologs can be in one-to-one relationships (HuA - MoA), many-to-one (HuA & HuB - ChC), or many-to-many (no examples in this tree, but would occur if there were a duplication in chicken). All of these relationships are identified by OrthoFinder.
Why Orthogroups?
There are several reasons why Orthogroups are the relevant way of analysing orthology relationships between species:
-
• Orthogroups allow you to analyse all of your dataAll of the genes in an orthogroup are descended from a single ancestral gene. Thus, all the genes in an orthogroup started out with the same sequence and function. As gene duplication and loss occur frequently in evolution, one-to-one orthologs are rare and limitation of analyses to on-to-one orthologs limits an analysis to a small fraction of the available data. By analysing orthogroups you can analyse all of your data.
-
• Orthogroups allow you to define the unit of comparisonIt is important to note that with orthogroups you choose where to define the limits of the unit of comparison. For example, if you just chose to analyse human and mouse in the above figure then you would have two orthogroups.
-
• Orthogroups are the only way to identify orthologsOrthology is defined by phylogeny. It is not definable by amino acid content, codon bias, GC content or other measures of sequence similarity. Methods that use such scores to define orthologs in the absence of phylogeny can only provide guesses. The only way to be sure that the orthology assignment is correct is by conducting a phylogenetic reconstruction of all genes descended from a single gene the last common ancestor of the species under consideration. This set of genes is an orthogroup. Thus, the only way to define orthology is by analysing orthogroups.
For a comprehensive overview of orthology and the OrthoFinder approach, you can watch David Emms’ conference talk on YouTube, from the 2020 symposium on Phylogenomics and Comparative genomics.